File Meta Sink
Write stream to file with meta-data headers.
These files represent data as binary information in between meta-data headers. The headers contain information about the type of data and properties of the data in the next segment of samples. The information includes:
- rx_rate (double): sample rate of data.
- rx_time (uint64_t, double): time stamp of first sample in segment.
- size (uint32_t): item size in bytes.
- type (::gr_file_types as int32_t): data type.
- cplx (bool): Is data complex?
- strt (uint64_t): Starting byte of data in this segment.
- bytes (uint64_t): Size in bytes of data in this segment.
Tags can be sent to the file to update the information, which will create a new header. Headers are found by searching from the first header (at position 0 in the file) and reading where the data segment starts plus the data segment size. Following will either be a new header or EOF.
Use of the pmt package to specify parameter Extra Dict requires an Import block with import pmt.
Parameters
(R): Run-time adjustable
- File (R)
- Name of file to write data to.
- Sample Rate
- Sample rate of data. If sample rate will be set by a tag, such as rx_tag from a UHD source, this is basically ignored.
- Relative_rate
- Rate change from source of sample rate tag to sink.
- Max Seg. size
- Length of a single segment before the header is repeated (in items).
- Extra dict.
- A PMT dictionary of extra information. This field is required, and pre-filled as
pmt.make_dict().
- Detached header
- Set to true to store the header info in a separate file (named filename.hdr)
- Unbuffered (R)
- If new samples are flushed to file at the end of each call to the work function
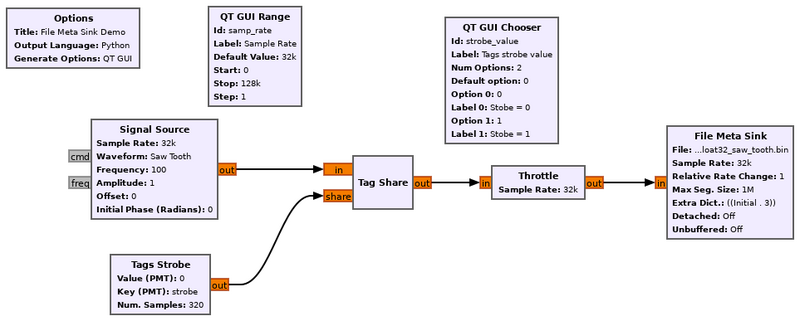
Example Flowgraph
In this example, the file meta sink block is saving both a data stream from the signal source generating a saw tooth waveform and stream tags that align with the start for the saw tooth waveform.
Source Files
- Header files
- [3]
- Public header files
- [4]
- Block definition
- [5]
FAQ
How is the file organized when it contains an embedded header?
A file with an embedded header is created by the file meta sink block is organized into multiple data segments that are sequential. Assume the waveform (Fig. 1.A) is being recorded to file. The waveform (Fig. 1.B) is converted to a data stream which may be annotated with stream tags [6]. In this example the stream tags indicate both the zero crossings (Fig. 1.D) and the positive peaks (Fig. 1.E) of the waveform. Each data segment in the file (Fig. 1.F) is composed of a header (Fig. 1.G) and the waveform data (Fig. 1.H). The header can be further decomposed into metadata and stream tags (Fig. 1.I). The metadata consists of information such as a sample rate, absolute position in the stream sequence and data type. The stream tags can indicate any useful feature or information about the data stream. A new data segment is created when either:
- A stream tag has been written (segments 1,2,3,5, and 6)
- The data segment size maximum has been reached which is specified by the parameter "Max Seg. size" (segment 4). In this segment no stream tags are recorded.
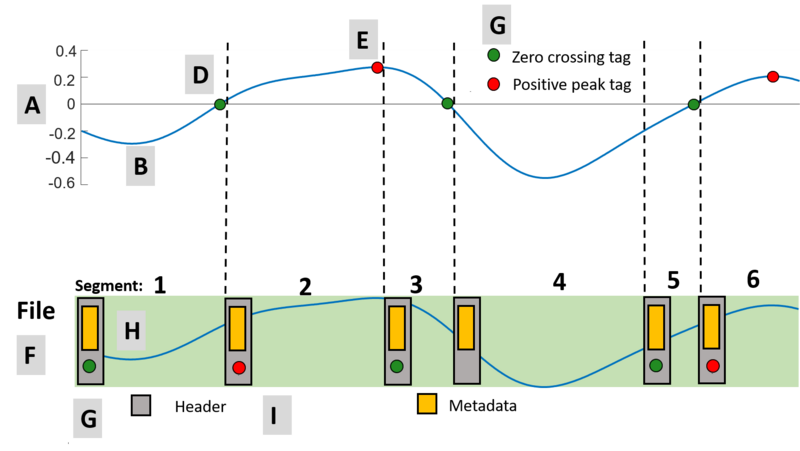
Figure 1: Recording a waveform to a single file with stream tags
How is the file organized when the header is detached?
The waveform (Fig. 2.A) is saved in a similar manner as with the embedded header except the data stream (Fig. 2.B) and header information (Fig. 2.C) are stored in separate files.
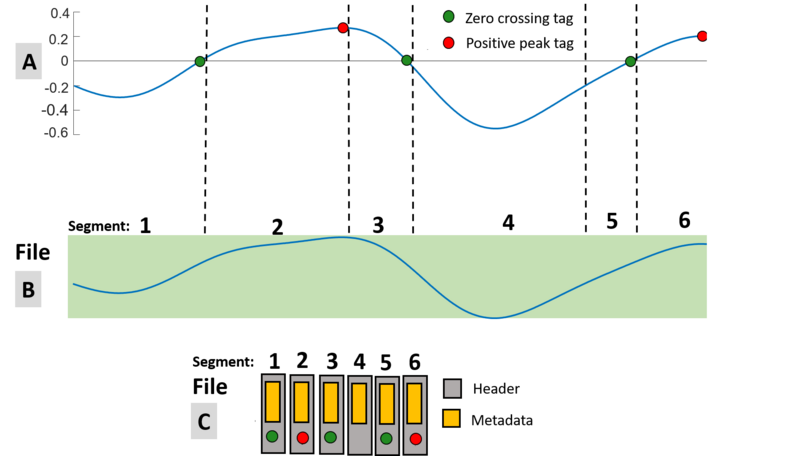
Figure 2: Recording a waveform to a data stream file and detached header file
How do I read a file with embedded headers in Python?
To read a file with embedded headers use the parse_file_metadata class provided by GNU Radio. The code below shows how to read the header including stream tags and how to read the data segment and convert it to a numpy array. This example does not handle the exceptions which may be thrown such as RuntimeError.
from gnuradio import gr,blocks
import pmt
import sys
from gnuradio.blocks import parse_file_metadata
import numpy as np
filename ='./data/data_float32_saw_tooth.bin'
max_data_segments_to_read = 3
print_output = True
fh = open(filename, "rb")
for ii in range(max_data_segments_to_read):
header_str = fh.read(parse_file_metadata.HEADER_LENGTH)
header = pmt.deserialize_str(header_str)
print(f"\n===Data segment {ii} ===")
header_info = parse_file_metadata.parse_header(header, print_output)
if(header_info["extra_len"] > 0):
extra_str = fh.read(header_info["extra_len"])
if(len(extra_str) != 0):
extra = pmt.deserialize_str(extra_str)
extra_info = parse_file_metadata.parse_extra_dict(extra, header_info, print_output)
data=np.fromfile(file=fh, dtype=np.float32, count=int(header_info['nitems']), sep='', offset=0)
print(f"{len(data)} data elements read")
fh.close()
The output from the above code for one of the data segments is shown below. The file was generated from the example flowgraph. The debug stream tag "strobe" and the key value pair Initial:3 which was in the Extra dict. parameter are shown.
...
===Data segment 2 ===
Version Number: 0
Sample Rate: 32000.00 sps
Seconds: 0.000000
Item size: 4
Data Type: float (5)
Complex? False
Header Length: 181 bytes
Extra Length: 32
Extra Header? True
Size of Data: 1280 bytes
320.0 items
strobe: 0
Initial: 3
320 data elements read
===Data segment 3 ===
....
What are examples of Extra dict. parameter?
The PMT values specified in the Extra dict. parameter will be included in every data segments' header. If this parameter is not needed then it should be initialized to pmt.make_dict(). Two examples of key value pair initialization are:
- If a single key value pair of 'Initial':3 is desired then the parameter could be
pmt.dict_add(pmt.make_dict(), pmt.intern('Initial'), pmt.from_long(3)). - If multiple key value pairs are desired, for example 'Initial':3 and 'Second':'value' is desired then the parameter could be
pmt.dict_add(pmt.dict_add(pmt.make_dict(), pmt.intern('Initial'), pmt.from_long(3)), pmt.intern('Second'), pmt.intern('value')).
An explanation of using PMT values in Python is described here [7]